Although the goal of gene therapy in preclinical models is rather focused on the impact of the protein encoded by the delivered gene on phenotypic outcome in disease models, our aim must be understanding the effects of this change in protein expression on the overall molecular pathways involved to define quantitative and predictive biomarkers.
This reflects a shift from a 1-gene-per-disease mindset to understanding of complex multigene disease processes, where phenotypic outcome is dependent on the behavior of interrelated networks and systems as a whole. Systems biology, including high-throughput experimental and computational studies that account for the complexity of host–disease therapy interactions, holds significant promise in aiding the development and optimization of gene therapy. With respect to target selection, a particular strength of systems biology studies is the ability to predict effects on the system that are not directly an intended target of the therapy and thereby identify hidden drivers of therapeutic success or failure. The idea behind this approach is that activity within a signaling network is probably more important than merely altered expression of a specific protein.

Typically, gene delivery studies use elevated levels of the therapeutic protein as confirmation that the treatment is working. However, the expression of the encoded protein is only the start of a process of the tissue responding to the intervention, and the protein will interact with many endogenous molecules and cause multifocal effects. It is the impact of these effects on the desired therapeutic outcome that may define the efficacy of the treatment and enable development of predictive biomarkers as companion diagnostics. Systems biology can aid in predicting changes and effects by incorporating dynamic information on the expressed protein and the overall network in which it is involved to deliver a holistic understanding of its mechanism of action and therapeutic efficacy. Potential approaches could pursue, for example, serial assessment of changes in the transcriptome (also including regulatory RNAs) of a target cell corresponding to the treatment over time. As already shown in other disciplines, in silico simulation can then integrate the time-resolved information and provide a computational model of the regulatory gene network that shapes the disease phenotype.
Within this framework, therapeutic factors can be considered as beneficial system perturbations, which is key to understanding how a particular pathway reacts. Thus, systems biology could assist in evaluating success and consequences of gene therapy and provide a bridge between therapeutic design and outcome. In addition, side effects and interactions of gene therapy with other drugs would also be identifiable and this could enable us to design around them. However, the systematic use of this approach is still underused relative to its potential.
Source ahajournals.org
DUC TIN Surgical Clinic
Tin tức liên quan
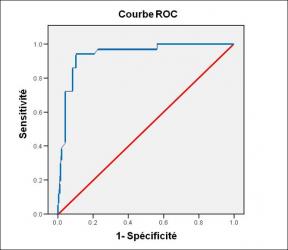
Performance diagnostique de l’interféron gamma dans l’identification de l’origine tuberculeuse des pleurésies exsudatives
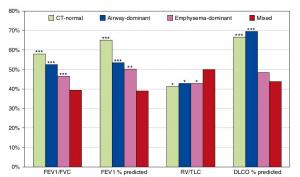
A Mixed Phenotype of Airway Wall Thickening and Emphysema Is Associated with Dyspnea and Hospitalization for Chronic Obstructive Pulmonary Disease.
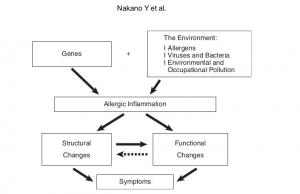
Radiological Approach to Asthma and COPD-The Role of Computed Tomography.

Significant annual cost savings found with UrgoStart in UK and Germany
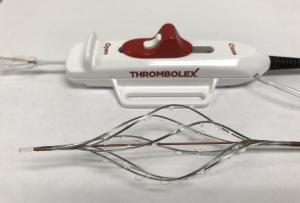
Thrombolex announces 510(k) clearance of Bashir catheter systems for thromboembolic disorders
Phone: (028) 3981 2678
Mobile: 0903 839 878 - 0909 384 389